Norine REST services
Description
Norine provides usefull REST[1] services to access the Norine database. Whereas classical web approaches are mainly dedicated to humans by providing rich and interactive interfaces (such as web forms or pie diagrams) for querying the database, RESTful service main objective is to allow integration with computers through software, applications or programs. This section describes such services that are provided by Norine and how to use them and finally integrate with your application. Most of the time, results are proposed in various output formats such as HTML, XML or Json.
Table of contents
- Peptides
- Monomers
- Links to external databases (PDB, PubChem, ...)
- Organisms
- Connect your software/database with Norine!
- Use the Smiles2Monomers tool as a web service
- Activity prediction web service
Peptides
Retrieve peptide details from a Norine ID
To retrieve details of a peptide (name, biological activity, family, structure...) from a Norine ID, simply request the following URI using a browser, an HTTP client tool (e.g. wget, curl, etc.) :
- http://norine.univ-lille.fr/norine/rest/id/<id_value>
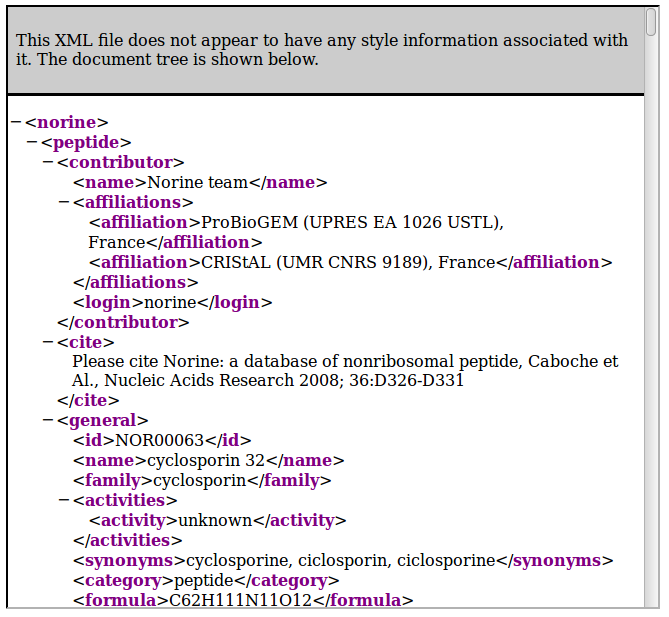
Example : http://norine.univ-lille.fr/norine/rest/id/NOR00123
By default, the server returns HTML content, as illustrated on the following figure:
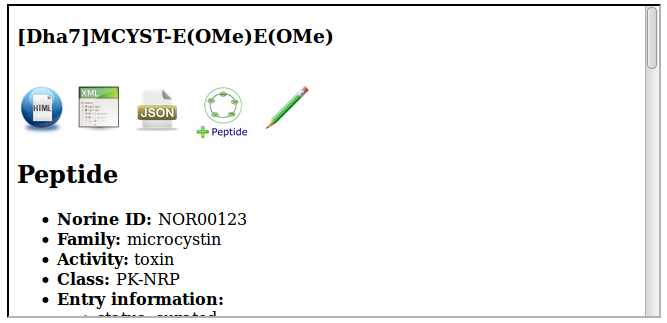
To get XML content, just add "/xml" to the path:
- http://norine.univ-lille.fr/norine/rest/id/xml/<id_value>
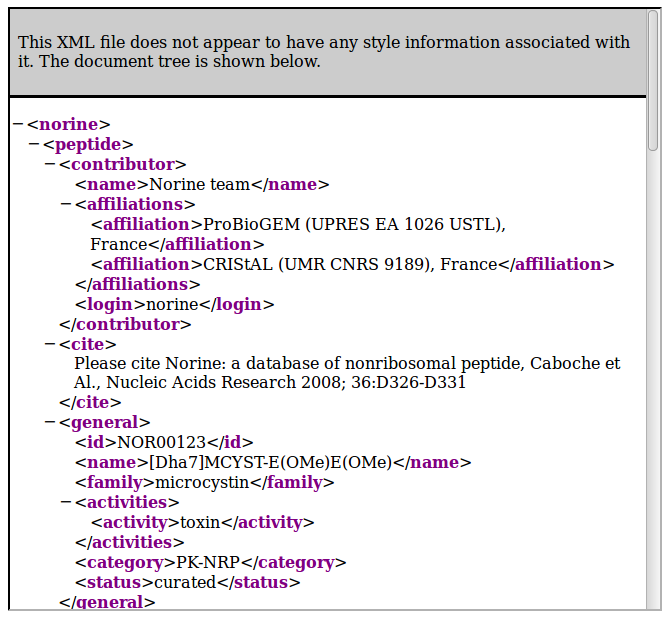
- http://norine.univ-lille.fr/norine/rest/id/json/<id_value>
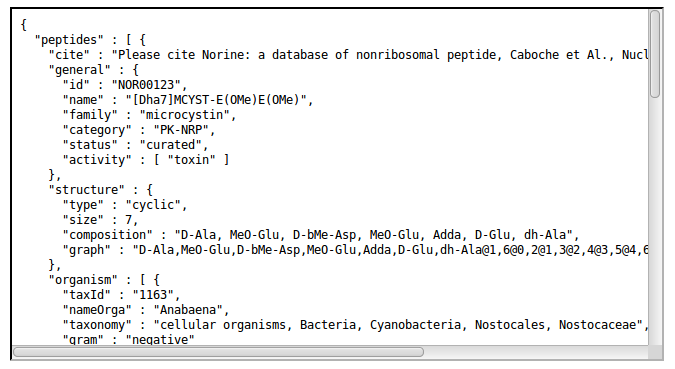
Retrieve peptide details by name (general search)
The general resource URI is the following:- http://norine.univ-lille.fr/norine/rest/name/<name_value>
This example gets all the peptides which name match the word cyclosporin. By default, the server returns HTML content, as illustrated on the following figure:
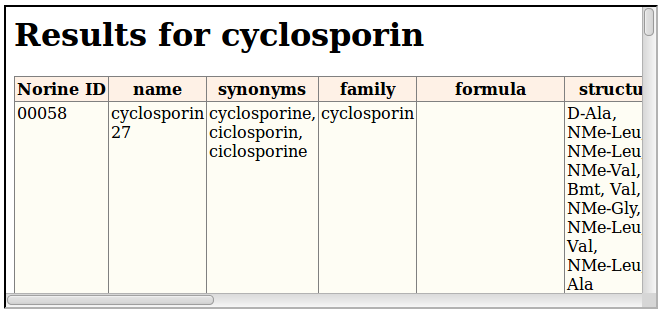
To get XML content, just add /xml to the URI, before the name value:
- http://norine.univ-lille.fr/norine/rest/name/xml/<name_value>
For Json, use:
- http://norine.univ-lille.fr/norine/rest/name/json/<name_value>
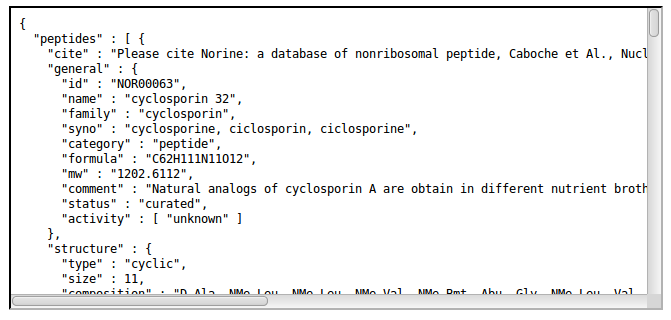
Get all peptides with smiles (JSON)
The general resource URI is the following:- http://norine.univ-lille.fr/norine/rest/peptides/json/smiles
Monomers
Retrieve all monomers
The general resource URI is the following:- http://norine.univ-lille.fr/norine/rest/monomers
This example gets all the monomers. By default, the server returns HTML content, as illustrated on the following figure:
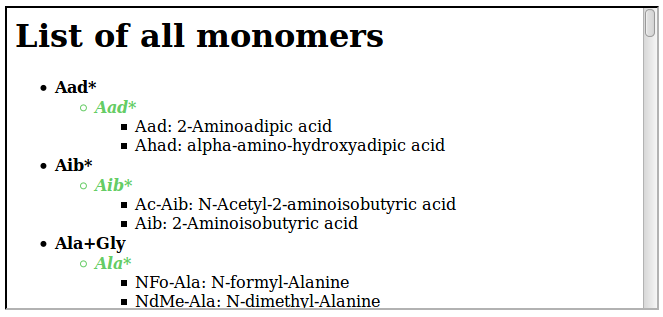
To get XML content, just add /hierarchicall/xml to the URI:
- http://norine.univ-lille.fr/norine/rest/monomers/hierarchical/xml
- http://norine.univ-lille.fr/norine/rest/monomers/flat/xml
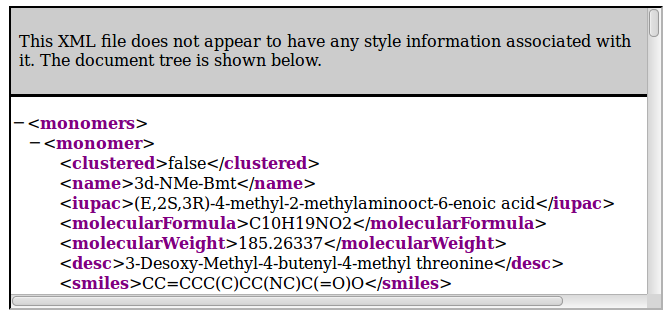
For Json, use:
- http://norine.univ-lille.fr/norine/monomers/hierarchical/json
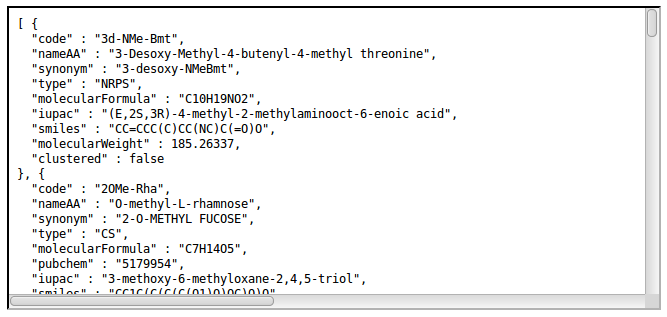
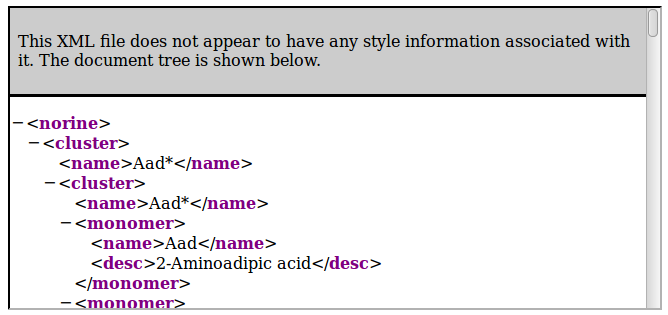
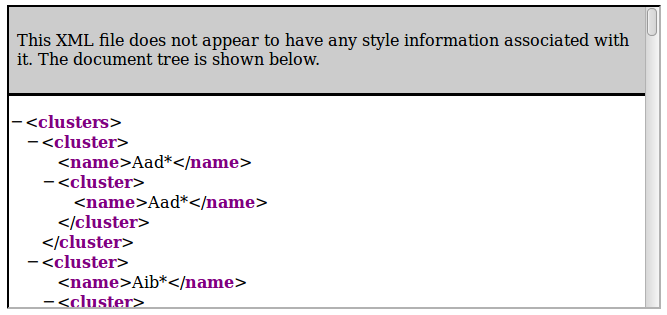
Retrieve clusters tree
The general resource URI is the following:- http://norine.univ-lille.fr/norine/rest/monomers/clusters/xml
Links to external databases (PDB, PubChem, ...)
Retrieve all externals access codes
The general resource URI is the following:- http://norine.univ-lille.fr/norine/rest/link/code/all
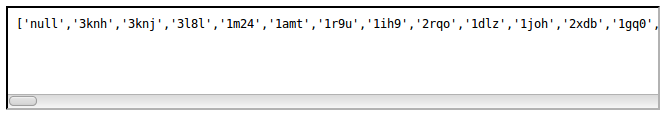
Retrieve database name from access codes
The general resource URI is the following:- http://norine.univ-lille.fr/norine/rest/code/<code_value>
Organisms
Retrieve organism details by name
The general resource URI is the following:- http://norine.univ-lille.fr/norine/rest/organism/xml/<organism_name>
For Json, use:
- http://norine.univ-lille.fr/norine/rest/organism/json/<organism_name>
Connect your software/database with Norine!
General search using an HTML form
The general search using a form has the same goal of previous examples but allows to combine multiple criterias
Using your favorite programming language
There are differents ways to connect your software to Norine web services. For instance, you can programmatically access the provided rest services by writing your own code in any programming language. Indeed, most of them provides libraries to create HTTP requests.The following example of code, written in Java language, simply request peptide details for ID#000123. This example use the standard Http client library provided by the Apache foundation.
Use the Smiles2Monomers tool as a web service
Getting monomeric composition and 2D graph from smiles
The general resource URI is the following:- http://norine.univ-lille.fr/norine/rest/s2m/graph/<smiles>
Amv,Hiv,D-Ala,dh-Ala@3,1@0,2@1,3@0,2
Retrieve all information on monomers
This allows to retrieve all information on monomers that compose the entered peptide such as the code, the complete name, the smiles of the monomer, the molecular formula and the molecular weight.The general resource URI is the following:- http://norine.univ-lille.fr/norine/rest/s2m/graph/json/<smiles>

Activity prediction
Biological activity prediction from graph and category
This service is based on the tool. The general resource URI is the following:
tool. The general resource URI is the following:- http://norine.univ-lille.fr/norine/rest/prediction?graph=<graph>[&category=<category>]
http://norine.univ-lille.fr/norine/rest/prediction?graph=Gly,Tyr,dhAbu,Thr,Thr,C14:0-OH(3),dhAbu,Gln,Thr,His @4,7 @6,3 @5,8 @1,8 @0,9 @2 @1,7 @0,6 @2,3,9 @4,8&category=lipopeptide will return the following result :
antimicrobial
If category is ommitted in the service URL pattern (optional), it will be considered by default as "pure peptide". The list of available categories is : pure peptide | peptaibol | glycopeptide | lipopeptide | lipopeptide with carbohydrate | chromopeptide | polyketide-NRP hybrid | unknown.[1] REST: Representational state transfer